go to: Ubiquitin System Products Dashboard
Ubiquigent Product Categories
New Products
Phospho Ubiquitins
E1 Activating Enzymes
E2 Conjugating Enzymes
E3 Ligases
Ubiquitin Binding Proteins
Deconjugating Enzymes (DUBs)
Ubiquitin/Ubl Modifiers
Proteasome
Kinases
Cell Lysates
Antibodies
HTS Assay Kits
Kits
Proteasome
Substrates
DUB Substrates
Ubiquitin Chains
- Why To Use Ubiquigent Reagents
● Fully annotated protein sequence
● Gradient SDS-PAGE purity analysis
● Tandem mass-spectrometric confirmation of identity
● Activity determination in application-specific assays
● Background literature synopsis provided for all products
● Standard formulations and fixed concentrations
● Online datasheets
● Expert technical support - Ubiquigent Application Note
Validation and Experimental Utility of the E2scan™ Kit version 2
"Linking Ubiquitin Research to Drug Discovery"
Ubiquigent Reagent Product Dashboard

About Ubiquigent Ltd.
Ubiquigent Limited is a highly focused provider of expertly produced reagent products and state-of-the-art drug discovery assay services surrounding the ubiquitin and ubiquitin-like signaling systems. Owned in part by the UK Medical Research Council (MRC) and the University of Dundee, Ubiquigent's state-of-the-art laboratory facilities are located adjacent to the MRC Protein Phosphorylation Unit and the Protein Ubiquitylation Unit the at the University of Dundee, Scotland, UK. Both of these research units were founded by renowned cell signaling scientist Professor Sir Philip Cohen, whose current interests include the interplay between protein ubiquitinylation and protein phosphorylation.
Ubiquigent's proximity and relationships provide it with ready access to cutting edge scientific information and technical expertise. In concert with it's own own expert staff, these elements allow Ubiquigent to offer not only the finest ubiquitin-related reagent products available today, but also assay and assay development services designed and conducted with an understanding of your needs that few companies can match.
Why To Choose Ubiquigent Reagents For Your Research Projects
A clear prerequisite for studying complex molecular systems is access to reagents of the highest integrity. Ubiquigent provides researchers with high integrity reagents characterized to give you confidence in your results.
All Ubiquigent reagents are characterized in house. Whenever possible, reagents are tested in application specific assays. Key features of Ubiquigent reagents are detailed on each batch-specific analytical data sheet.
●Fully annotated protein sequence
●Gradient SDS-PAGE purity analysis
●Tandem mass-spectrometric confirmation of identity
●Activity determination in application-specific assays
●Background literature synopsis provided for all products
●Standard formulations and fixed concentrations
●Data sheets available online; both current and historic
Overview of Protein Ubiquitylation
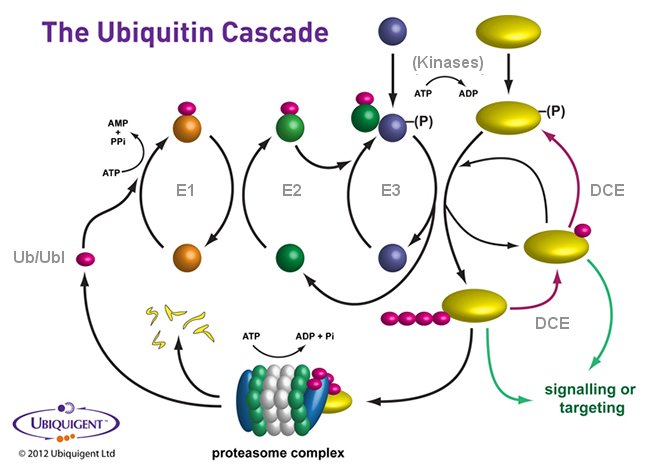
Ubiquitylation, like phosphorylation, describes a reversible post-translational protein modification. Ubiquitylation or 'ubiquitination' may control the protein substrate’s destiny – in respect of its turnover – or its signalling functionality. It is a process that refers to the covalent attachment of a small, 76 amino acid protein called ubiquitin to the epsilon-amino group of a lysine residue residing within a substrate protein – which may also be another ubiquitin molecule. This results in either mono- or poly-ubiquitylation of the substrate; the latter being where chains of ubiquitin are attached to the substrate protein. The structure of the chain determines whether a protein modulates a specific signalling cascade or may become degraded in a proteasomal or lysosomal dependent manner. Mono-ubiquitylated proteins may be further ubiquitylated to form polyubiquitin chains and deubiquitylases may act on either mono-ubiquitylated or poly-ubiquitylated substrates to remove the ubiquitin monomers or chains respectively.
The enzymes of the ubiquitylation pathway play a pivotal role in a number of cellular processes including, but not exclusively, the targeted proteasomal degradation of substrate proteins. Three classes of enzymes are involved in the process of substrate ubiquitylation; activating enzymes (E1s), conjugating enzymes (E2s) and protein ligases (E3s). Ubiquitylation of substrate proteins depends on the sequential action of these three enzymes. In an ATP-dependent first step, an E1 enzyme forms a thioester linkage with ubiquitin which is then transferred to the sulphydryl group of the active-site cysteine on an E2 enzyme forming a ubiquitin-thioester intermediate. An E3 then acts as an adaptor to bind both substrate protein and E2 'loaded' with ubiquitin. The E3 facilitates isopeptide bond formation between ubiquitin and the substrate protein.
Although still a post-translational modification, albeit involving a functional protein rather than a function group, ubiquitylation is a much more complex process than phosphorylation mainly due to the ability of ubiquitin to form polyubiquitin chains of a variety of different linkage types and complexity, but also because there are further related ubiquitin-like (UBL; including SUMO, NEDD8, ISG15, and FAT10) proteins that may each follow a similar specific enzymatic cascade but resulting in different outcomes for the UBL modified target substrate. At a further level of system complexity modifications of either the E3 ligase or the substrate may alter their ability to modify substrates or themselves be modified; for example by NEDDylation (NEDD8 being a further UBL) in the case of E3 ligases or phosphorylation in the case of both E3 ligases and substrates.
