DNase activity of Labiase
A previous questionnaire survey showed that labiase was most commonly used for the preparation of genomic and plasmid DNA from lactic acid bacteria. Thus, the DNA degradation activity of labiase was examined.
Methods
1. A 0.5% labiase solution was prepared using McIlvaine buffer (pH 4.0).
2. To 50 μl of the 0.5% labiase solution, 1 μg of DNA was added as a substrate.
3. Five reaction solutions were prepared. Of these, two each were heated at 30oC for 1 or 4 hours, and the remaining one was not heated as a blank (0 hours).
4. The above five samples were treated with phenol and chloroform twice, followed by ethanol precipitation and washing.
5. The precipitate was dried up and dissolved in 45 µl of TE. To the solution, 5 µl of 10x loading buffer was added for 0.8% agarose gel electrophoresis.
Lot Nos. JJAD1 and AAAC2 were tested.
Results
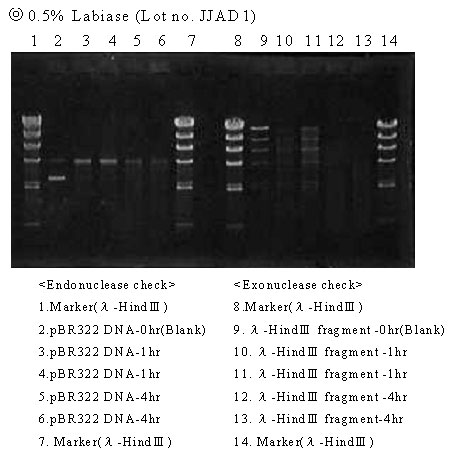
Results
Lot No. JJAD1: The band(s) of the blank disappeared for both endo- and exonucleases, suggesting the presence of DNase activity.
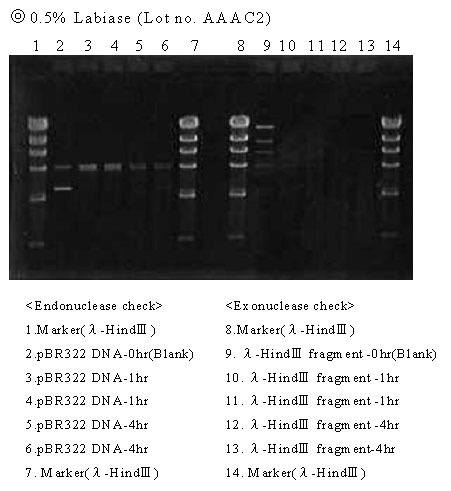
Results
Lot No. AAAC2: The band(s) of the blank also disappeared for both endo- and exonucleases, suggesting the presence of DNase activity.
Inhibition of the DNase activity of Labiase
Thus, the DNA degradation activity of labiase should be inhibited in preparing genomic and plasmid DNA from lactic acid bacteria using labiase. In the present study, whether it could be inhibited by a DNase inhibitor (EDTA) was examined.
Methods
1. A 0.5% labiase solution was prepared using McIlvaine buffer (pH 4.0).
2. To 45 μl of the 0.5% labiase solution, EDTA (or distilled water as a control) was added at a final concentration of 5, 25, or 50 mM. To the solution, 1 μg of DNA was added as a substrate.
3. Three reaction solutions were prepared. Of these, one was heated at 30oC for 1 hour, and another was heated for 4 hours. The remaining one was not heated as a blank (0 hours).
4. The above three samples were treated with phenol and chloroform twice, followed by ethanol precipitation and washing.
5. The precipitate was dried up and dissolved in 45 µl of TE. To the solution, 5 µl of 10X loading buffer was added for 0.8% agarose gel electrophoresis.
Only Lot No. AAAC2 was tested.
(Results) Results are shown with or without 5 mM EDTA.
These photos demonstrate that the band(s) present in the blank completely disappeared at 4 hours without EDTA and no DNA degradation was noted with 5 mM EDTA. Similar results were obtained at final concentrations of 25 and 50 mM. Thus, both DNase activities (endo- and exonucleases) in labiase were completely inhibited with 5 mM EDTA in the reaction solution.
Conclusion
The DNase (endo- and exonucleases) activities of labiase are inhibited and pose no procedural problem at an EDTA concentration for routine DNA handling.


