PUREfrex Product Pages
- PUREfrex 2.0
- (for protein synthesis)
- PUREfrex 2.1
(for proteins with disulfide bonds) - PUREfrex 1.0
(for basic research and mechanism studies) - PUREfrex 1.0 Custom
(formulated to your specs) - Reaction Supplements
- (EF-P, DnaK, GroE, DsbC Set, PDI Set)
PUREfrex Product Manual and Package Inserts
- PUREfrex 2.0/2.1 Kit Technical Manual
- 2.0/2.1 full version (34 pages)
- Package Inserts
- PUREfrex Kits
(reagent sets for Y ml rxn mix x number of reagent sets) - 2.0 - 100 uL
2.0 - 250 uL
2.0 - 2 mL
2.0 - 5 x 2mL
2.0 - 25 x 2mL
2.1 - 100uL
2.1 - 250uL
2.1 - 2mL
2.1 - 5 x 2mL
2.1 - 25 x 2mL
1.0 - 250uL
1.0 - 2mL
1.0 - 5 x 2mL
1.0 - 25 x 2mL - PUREfrex Reaction Supplements
(reagent sets for Y ml rxn mix x number of reagent sets)
DnaK Mix - 500 uL
DnaK Mix - 5 x 2mL
DnaK Mix - 25 x 2mL
GroE Mix - 500uL
GroE Mix - 5 x 2mL
GroE Mix - 25 x 2mL
DsbC Set - 500uL
DsbC Set - 5 x 2mL
DsbC Set - 25 x 2mL
PDI Set - 500uL
PDI Set - 5 x 2mL
PDI Set - 25 x 2mL
EF-P - 500uL
EF-P - 5 x 2mL
EF-P - 25 x 2mL
- PUREfrex Kits
- Brochure

Download
Key Technical Notes
- ♦ PUREfrex Template Preparation
- ♦ Six Tips on PUREfrex DNA Template Design
- ♦ Synthesis of Protein Toxin (Gelonin) using DnaK Mix
- ♦ Sequence of DHFR DNA
- ♦ Influence of reducing agents on the redox state
- ♦ Improvement of specific activity by methylation of release factors (RF1, RF2).
- ♦ Improvement of the translation efficiency of consecutive proline residue-containing proteins by EF-P.
- ♦ Synthesis of proteins containing disulfide bonds with PDI and DsbC.
- ♦ Synthesis of functionally active proteins having disulfide bonds.
PUREfrex FAQ
PUREfrex in the Literature
Manufacturer's Site Links
- PUREfrex at Gene Frontier
- See All FAQ, Technical Notes, Posters, and Literature Citations at GeneFrontier Sites
Safety Data Sheets
- 2.0 SDS USA EU
- 2.1 SDS USA EU
- 1.0 SDS USA EU
- DnaK Mix USA EU
- GroE Mix USA EU
- DscC Set USA EU
- PDI Set USA EU
- EF-P USA EU
Posters
- General Protein Synthesis
- Improvement of translational efficiency by N-terminal codon optimization in the reconstituted cell-free protein synthesis system [2016]
- Improvement of the translation efficiency of proline residue-containing proteins using the reconstituted cell-free protein synthesis system (PUREfrex) [2018] VIEW
- Investigation on how to synthesize active proteins by using a reconstituted cell-free protein synthesis system (PUREfrex) [2020]
- Disulfide Bonding
- Synthesis of proteins containing disulfide bonds using a reconstituted cell-free protein synthesis system (PUREfrex). [2019]
- Synthesis of proteins containing disulfide bonds using PUREfrex® with human protein disulfide isomerase [2018] VIEW
- Synthesis of functionally active proteins containing disulfide bonds using the new PURE system (PUREfrex) [2011]
- Synthesis of Immunoglobulins and Biologics
- Unique Cell-Free Protein Synthesis System, PUREfrex -Useful Platform for Protein Expression in the Development of Biologics- [2018] VIEW
- Preparation of labeled cyclic peptides with PUREfrex [2017]
- Development of a method for the synthesis of aglycosylated full-length IgG using PUREfrex [2017]
- Efficient in vitro expression of aglycosylated full-length IgG using a reconstituted cell-free protein synthesis system, PUREfrex 2.0 [2016]
- In vitro production of Fab, Toxin, and Immunotoxin using PUREfrex®2.0 with increased productivity [2015]
- Synthesis of functionally active proteins containing disulfide bonds using the new PURE system (PUREfrex) [2011]
PUREfrex FAQ Page

state-of-the-art in vitro protein synthesis by
"Up to 100's of micrograms/ml in 240 minutes"Template DNA FAQ
Q1: What amount of the template DNA is added to the reaction mixture?
0.5-3 ng per 1 kbp DNA for 1 µL of the reaction mixture. The amount of DNA is calculated based on number of DNA molecules. For example, in case of 6 kbp of plasmid, regardless of length of ORF, 3 to 18 ng of DNA is added for 1 µL of the reaction mixture.
Q2: Can TE buffer be used for dissolving the template DNA?
No. Buffers containing EDTA should not be used for dissolving DNA, because EDTA inhibits transcription and translation reaction. PUREfrex®
Q3: What are key points to use PCR product as the template DNA?
We recommend that the template DNA is prepared by two-step PCR. An overview of a two-step PCR is illustrated below and primer sequences are given. Text sequence is also available.
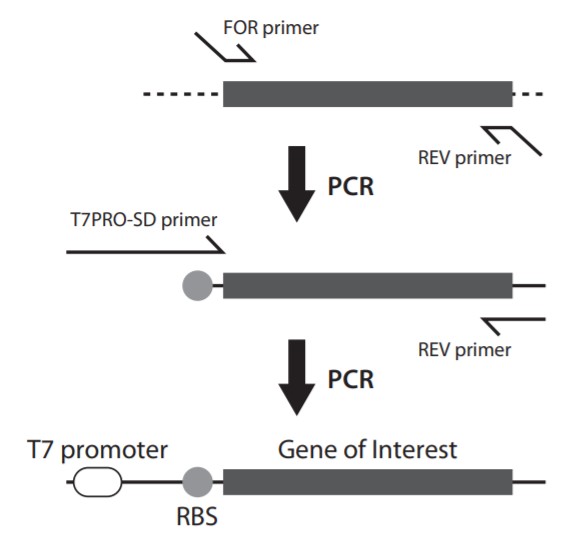

<p?Two- step PCR Primer purity and amount of PCR product should be a single band by electrophoresis. Otherwise unexpected polypeptides may be synthesized. If extra bands are observed despite optimizing PCR conditions, please purify the desired band by gel extraction. We recommend blue light for gel extraction, not UV light. UV light causes DNA damage which can lead to an unexpected transcription termination. Even using blue light, the band should be exiised quickly from the get.
PCR reaction mixture containing the amplified template DNA can be directly added to PUREfrex® reaction mixture. However, some components in the PCR reaction mixture inhibit transcription and/or translation reaction. To prevent carrying inhibitory components from the PCR reaction mixture, we recommend adding less than 0.1 volume of the PCR reaction mixture to PUREfrex® reaction mixture. If the concentration of the template DNA is not enough for protein synthesis, you may concentrate DNA solution with a purification kit or by gel extraction.
Q4: What are key points to use plasmid DNA as the template DNA?
To include essential elements T7 promotor, SD sequence, and T7 terminator are essential elements. We don’t recommend using vectors including Lac operator because it causes decrease on productivity depending on the target protein. RNase contamination RNase digests the transcribed RNA in the reaction mixture, resulting reduction of the productivity of the target protein. Many commercial kits can be used for purifying plasmid DNA. However, in some cases, RNase in the lysis buffer is brought over to the purified DNA fraction. If there is a possibility that RNase is contained in the purified plasmid DNA fraction, we recommend Phenol/Chloroform extraction followed by ethanol or isopropanol precipitation for removing RNase. Addition of RNase inhibitor to the PUREfrex® reaction mixture is also effective. PUREfrex®
Q5: Should I codon optimize my PUREfrex® template?
Yes. We recommend that you use the optimal codons for E. coli because the PUREfrex® derives from E. coli. Moreover, N-terminal codons (2nd-6th codon following 1st ATG) should be changed to AT-rich codon (not rather than the most frequently used codon) to increase the productivity. Please refer the results in our posters, which are partially described in Japanese but all results in English. “Improvement of translational efficiency by N-terminal codon optimization in the reconstituted cell-free protein synthesis system” (2016) »Poster1; Fab, His tag, etc. (0.7 MB) »Poster2; various proteins, membrane protein, etc. (2.5 MB)
Protein Synthesis FAQ
Can I synthesize eukaryotic proteins using the PUREfrex® kit?
Yes. The PUREfrex® is a reconstituted cell-free protein synthesis kit composed of E. coli ribosomes and translation factors, but eukaryotic proteins from such as mammalian and plant can also be synthesized. The productivity of the target protein may depend on the property of nucleotide sequence encoding the target protein, such as GC contents or frequency of rare codon.
Q2: How much can I synthesize the target protein using the PUREfrex® kit?
It depends on the target protein.
Dihydrofolate reductase from E.coli can be synthesized with about 600 μg per mL of the reaction.
Q3: Can I synthesized proteins of more than 100 kDa?
YES.
We have synthesized the protein of 116 kDa using PUREfrex®.
Q4: What are the recommended reaction conditions for PUREfrex®?
We recommend that PUREfrex® protein synthesis reactions are carried out at 37° for 2-4 hours.
Q5: Can I synthesize and purify a tagged protein?
YES.
In fact, with PUREfrex, you can use any tag because none of protein components of PUREfrex® are tagged, nor were they ever tagged for purification or detection. For example, a HIS-tagged synthesized target protein can be purified with metal-chelating resin by applying the reaction mixture directly.
Q6: Is synthesized protein modified by glycosylation or phosphorylation?
NO.
There is no post-translational modification because PUREfrex® is constituted only by translation factors.
Q7: Does PUREfrex® contain any molecular chaperones?
NO.
PUREfrex® does not contain any chaperones, but you can add chaperones, such as Hsp70. You may prepare chaperone by yourself or buy it from Cosmo Bio USA or another company. GeneFrontier offers molecular chaperones perfectly formatted for PUREfrex® reactions. Please click the "Reaction Supplement" links on the sidebar of this page for more information.
Q8: Can I synthesize a protein containing disulfide bonds with PUREfrex®?
YES.
DsbC Set (GFK-PF005-0.5-EX) and PDI Set (#GFK-PF006-05-EX) are PUREfrex® supplements for synthesizing proteins whose activity depends on disulfide bonding. The use of PUREfrex® 2.1 (#PF213-0.25-EX) is recommended to optimize the reaction conditions as it was developed for this purpose and allows for the empirical determination of optimal re-dox conditions.
Q9: Can I synthesize a membrane protein with PUREfrex®?
YES.
However, synthesized membrane proteins are likely to aggregates. To obtain a membrane protein inserted into a lipid bilayer, you may add a lipid bilayer such as liposomes to your PUREfrex® reaction.
Q10: Can I synthesize a radiolabeled protein with [35S] methionine or [3H] leucine?
YES.
A radiolabeled protein can be synthesized by adding radioisotope-labeled amino acids, such as [35S] methionine or [3H] leucine, to PUREfrex®. The amino acid content is 0.5 mM for PUREfrex®1.0 and 2 mM PUREfrex®2.0. Please consider these concentrations and optimize conditions for your labeling.
Q11: Can I use any promoter besides T7 promoter?
We recommend using template DNA with a T7 promoter because PUREfrex® contains T7 RNA polymerase for transcription. When you use other polymerases, please generate template DNA with a suitable promoter for the selected polymerase.
Q12: I can’t get DHFR using DHFR DNA in the kit as positive control.
A1: The kit may could have become inactivated. To avoid inactivation, please store kits and components as direct and divide into aliquots to reduce freeze-thaw cycles as much as possible.
A2: Your reactions may be contaminated by nuclease. To avoid nuclease contamination, please insure all your reagents and apparatus are nuclease-free.
Q13: I can get DHFR using the DHFR DNA template included with the kit, but I can’t get my protein of interest or only a very low amount.
- The kit may could have become inactivated. To avoid inactivation, please store kits and components as direct and divide into aliquots to reduce freeze-thaw cycles as much as possible.
- Your reactions may be contaminated by nuclease. To avoid nuclease contamination, please insure all your reagents and apparatus are nuclease-free. Plasmids are not infrequently contaminated with RNase from the plasmid preparation procedure. So, please confirm that RNase is removed from the plasmid.
- The construct of template DNA may be incorrect. It is necessary that the template contain a T7 promoter sequence, a ribosome binding site, initiation codon, and stop codon for the template DNA for PUREfrex®. Please see the Template FAQ above and your kit instructions.
- Secondary structure of transcripts may prevent the translation reaction. In this case, please optimize the template sequence to solve the problem of secondary structure.
