(CLICK FOR ORDERING INFORMATION)
(CLICK FOR COMPREHENSIVE ANNOTATED BIBLIOGRAPHY)
PUREfrex® is a reconstituted coupled transcription/translation system based on the PURE system originally developed by the Ueda group (Shimizu et al., 2001) and later commercialized as PURESYSTEM® (Shimizu and Ueda, 2010) by BioComber Co. Untagged protein components permit addition of any desired tag (including His-tags) to a protein of interest. For template generation we recommend Protein Express Linear Template PCR kits (available in two versions: His-tag only or ProX-tag + His-tag) or expression plasmids from Protein Express (pROX-FL92.1: ProX-tag + His-tag) or BioComber (pURE1-4: no tags).
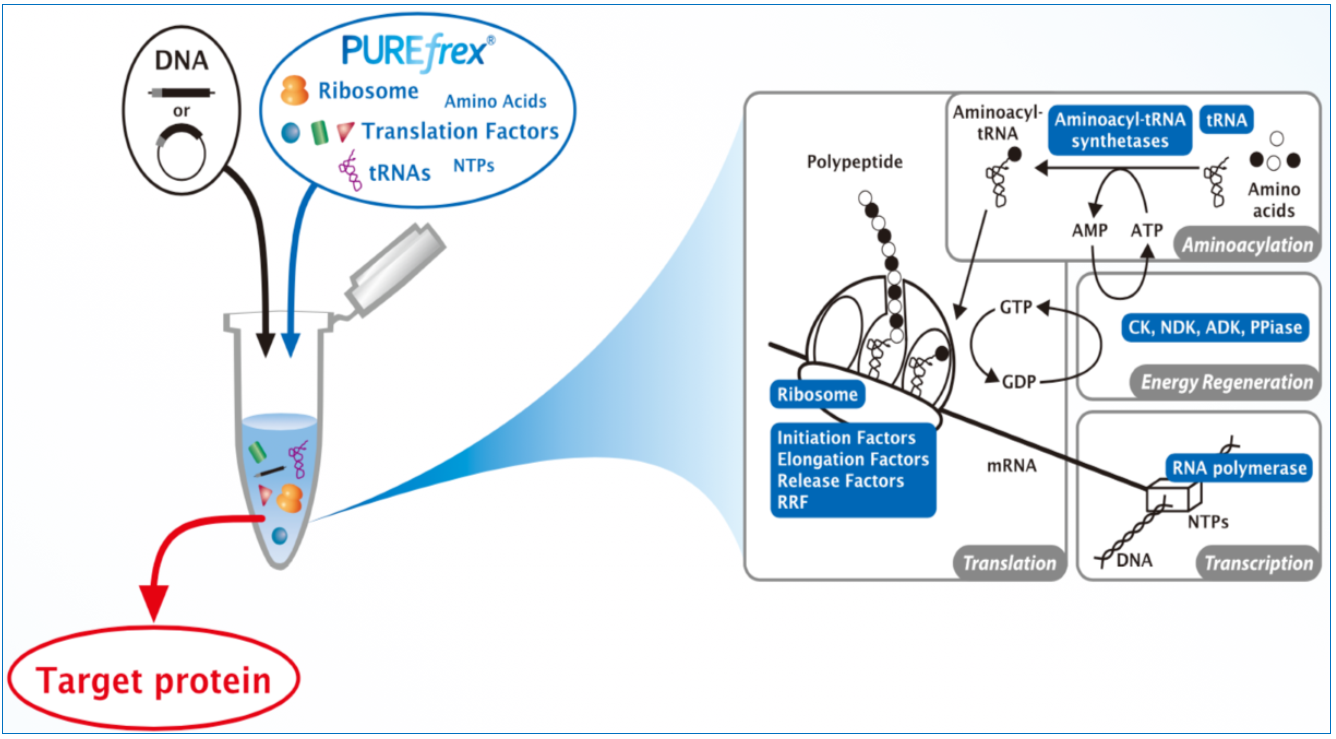
All necessary macromolecular components needed for in vitro transcription and translation are highly purified from E. coli and supplied as two solution mixes (A and B). The system includes modules for aminoacylation, energy regeneration, transcription and translation.
The advantages of PUREfrex® include reduced levels of contaminating proteases, nucleases, and phosphatases, beta-galactosidase and LPS. Compared to cell lysate-based systems, the more defined and modular chemistry of PUREfrex® leads to greater yield, reproducibility and flexibility. PUREfrex® is available in two forms: PUREfrex® 2.0 and PUREfrex® 2.1. PUREfrex® 2.1 is identical to PUREfrex® 2.0 with the added ability for the user to control redox conditions; this is useful for proteins that require special redox conditions for proper folding and disulfide bond formation.
PUREfrex® 2.0 and PUREfrex® 2.1 contain no molecular chaperones. When proper folding, activity and solubility of your protein of interest requires molecular chaperones, Gene Frontier offers two molecular chaperone supplements for use with PUREfrex® 2.0 and PUREfrex® 2.1: GroE Mix and DnaK Mix. GroE Mix comprises an optimized mixture of highly purified E. coli GroEL (Hsp60) and GroES. DnaK Mix comprises an optimized mixture of highly purified E. coli DnaK, DnaJ and GrpE. DnaK (Hsp70) has ATPase activity and is stimulated by co-chaperones DnaJ and GrpE. DnaJ facilitates the ATPase activity of DnaK and can bind to hydrophobic protein regions. GrpE stimulates ADP/ATP exchange of DnaK. For a given protein the relative suitability of GroE Mix and DnaK Mix for promoting folding, solubility and activity must be determined empirically.
Gene Frontier also offers two additional supplements to promote proper disulfide bond formation: DS Supplement (soon to be phased out and replaced with DsbC Set) and PDI Set. Formation of disulfide bonds is important for the folding, stability and activity of many proteins, including cell surface receptors and secreted proteins. Disulfide bonds are formed by oxidation of pairs of cysteine sulfhydryl (SH-) groups. Therefore, an oxidizing environment is necessary for disulfide bond formation. Additionally, the correct pairing of cysteines is promoted by Disulfide bond isomerase (e.g. E. coli DsbC) which can catalyze disulfide bond exchange reactions. DS supplement comprises highly purified E. coli DsbC and oxidized Glutathione (CSSG). PDI Set comprises oxidized glutathione (GSSG), human PDI (Protein disulfide isomerase) and human Ero1α (ER oxidoreductin-1 to reoxidize PDI).
Benefits of PUREfrex®
- PUREfrex® supplements expand the potential to express properly folded, disulfide-bonded, functional proteins.
- Molecular chaperones - Assists folding and improves solubility of proteins that require molecular chaperones for proper folding.
- DnaK Mix – DnaK Mix comprises highly purified and ratio-optimized E. coli DnaK, DnaJ and GrpE. DnaK, known as Hsp70, has ATPase activity and is stimulated by co-chaperones DnaJ and GrpE. DnaJ facilitates DnaK ATPase activity and binds to hydrophobic regions of synthesized proteins. GrpE stimulates ADP/ATP exchange of DnaK. DnaK Mix works well with PUREfrex® series and DsbC Set.
- GroE Mix - GroE Mix comprises highly purified and ratio-optimized E. coli GroEL (known as Hsp60) and GroES (working in conjunction with GroEL). GroE Mix works well with PUREfrex® series.
- Disulfide bond enhancers - Formation of disulfide bonds is important for folding and stability of secretory proteins such as enzymes or antibodies. Disulfide bonds are usually formed by oxidation of sulfhydryl groups (SH-) of adjacent cysteine residues. Therefore, disulfide bond formation efficiency depends on redox state. Additionally, disulfide bond isomerase which can catalyze the exchange of disulfide bridges may be required for correct cysteine pairing.
- DsbC Set - DsbC Set comprises highly purified E. coli DsbC (a disulfide bond isomerase) which can catalyze disulfide bridge exchange, and GSSG (oxidized Glutathione) to enforce an oxidized environment.
- PDI Set - PDI Set comprises oxidized glutathione (GSSG), human PDI (protein disulfide isomerase) and human Ero1α (ER oxidoreductin-1 to reoxidize PDI).
- Rapid synthesis of:
- Membrane proteins
- Antibodies
- Antigens
- Cytokines
- Metal cofactor-dependent enzymes
- Phosphoproteins
- Antimicrobial peptides
- Therapeutic peptides
- All proteinaceous components of PUREfrex® are tag-free, permitting users to choose any desired tag for purification and monitoring.
- Extremely low contamination from LPS, proteases, nucleases, β-galactosidase and phosphatases
- Free of metabolic side reactions (found in cell extracts) that deplete amino acid pools
- Flexibility of a modular system
- Greater reproducibility resulting from more defined chemistry
- Cleaner resolution of expressed products due to elimination of background proteins found in traditional cell extracts
- Permits manipulation of protein synthesis reaction conditions, reactants, and direct monitoring of performance
- Rapid production of ‘difficult’ proteins that may be toxic to conventional cell protein factories
- Permits protein synthesis from different templates (DNA: circular plasmids, linear amplicons, rolling circle amplification; RNA: mRNAs)
- High-throughput expression of multi-protein complexes
- Generation of freeze-dryable point of use nanoscale biosensors:
- Biosensors for endocrine disruptors
- Biosensors for viruses
- Biosensors for small molecules
- Biosensors for quorum sensing molecules
- Biosensor for histamine
- Tetramerization-driven biosensor for anti-tag antibodies
- High throughput functional genomics/proteomics
- mRNA display
- Ribosome display
- Site-specific protein labelling/mutagenesis with non-natural amino acids. Cellular systems are susceptible to toxic side-effects of non-natural amino acids in cellular proteins or recombinant products; cell-free systems can be easily tailored to accommodate unusual side chain structures by modifying components of the translation machinery; for example, mutation of EF-Tu has been shown to improve the incorporation efficiency of non-natural amino acids with bulky side chains (Doi et al. 2007):
- Biotin labeling
- PEGylation
- Post translational modification
- Fluorescence labeling for molecular imaging
- Double fluorescent labeling for single molecule FRET
- Protein structure/function analysis
- Stable isotopic labeling of internal standards for targeted quantitative proteomics (when proteins are expressed in living cells or cell extracts, labeling is complicated by metabolic processes that dilute isotope or scramble labeling patterns as a result of molecular transformations)
- In vitro protein evolution
- Rapid drug candidate screening
- Rapid gene construct screening
- Rapid confirmation of coding potential of ORFs
- Study of artificial cells
- Study of protein translation
- Study of protein folding
- Study of protein engineering
- Study of ribosome function
- Study of ribosome stalling
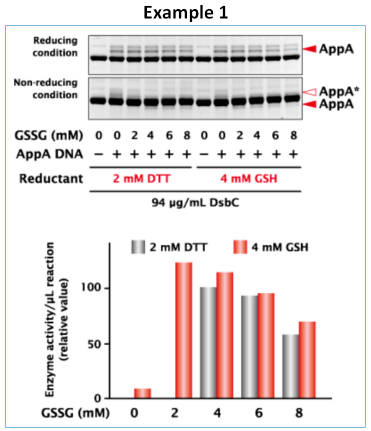
Synthesis of E.coli phosphatase (AppA)
E. coli phosphatase (AppA) was synthesized with PUREfrex® 2.1 and DS supplement at 37°C for 4 hours. Protein yield was similar under different redox states, but protein activity was highest with 4 mM GSH and 2 mM GSSG (oxidized glutathione).

Synthesis of IgG
Immunoglobulin G (IgG) is a Y-shaped tetramer comprising two heavy chains (HC) and two light chains (LC) joined by disulfide bonds. HCs and LCs were synthesized in a single tube with PUREfrex® 2.1, DS supplement and DnaK Mix at 37°C for 16 hours. Although the yields of HCs and LCs were similar across all conditions, the yield of tetrameric IgG was highly dependent on the nature and amount of reducing agent.
Annotated Cell-free Transcription/Translation Systems Bibliography
The ability to reconstitute protein synthesis from well-defined, purified components (the PURE system), has transformed the cell-free protein synthesis approach from a specialized analytical tool to a powerful preparative method with broad applicability. Currently, there are two major commercial sources of PURE system kits and ancillary supplements: PUREfrex from Gene Frontier and PURExpress from New England Biolabs; a smaller vendor (BioComber) also produces a commercial version of the PURE system called PURESYSTEM. Below are annotated bibliographies highlighting the many applications of this powerful technology, divided into sections for: PUREfrex®, PURExpress®, PURESYSTEM® and general cell-free protein synthesis.
Articles citing PUREfrex®
Use of PUREfrex® to demonstrate that stable translational arrest of nascent bacterial SecM protein requires an interaction with the ribosomal outer surface. SecM containing the non-natural photo-crosslinking amino acid Bpa (p-benzoyl-L-phenylalanine) was produced using a PUREfrex® kit lacking RF1. UV-triggered crosslinking revealed nascent SecM protein interactions with ribosomal protein L23. (Muta, M., Iizuka, R., Niwa, T., Guo, Y., Taguchi, H., Funatsu, T. (2020). Nascent SecM chain interacts with outer ribosomal surface to stabilize translation arrest Biochemical Journal 477(2), 557-566. https://dx.doi.org/10.1042/bcj20190723)
A review describing newly discovered CysSSH synthase (CPERS) activity of E. coli cysteinyl-tRNA synthetase (EcCARS) and mammalian CARS2. PUREfrex® is used to confirm the intact protein synthesis of mutant versions of EcCARS. (Fujii, S., Sawa, T., Motohashi, H., Akaike, T. (2019). Persulfide synthases that are functionally coupled with translation mediate sulfur respiration in mammalian cells British Journal of Pharmacology 176(4), 607-615. https://dx.doi.org/10.1111/bph.14356)
Use of PUREfrex® to generate a protocell-based biosensor system. Giant unilamellar vesicles (GUVs) containing PUREfrex® and suitable templates expressed hybrid membrane fusion proteins comprising extracellular tags, a transmembrane domain and a dimeric inactive mutant of beta-glucuronidase (GUS) that regains catalytic activity upon anti-tag antibody-dependent tetramerization. (Su, J., Kitaguchi, T., Ohmuro-Matsuyama, Y., Seah, T., Ghadessy, F., Hoon, S., Ueda, H. (2019). Transmembrane signaling on a protocell: Creation of receptor-enzyme chimeras for immunodetection of specific antibodies and antigens. Scientific reports 9(1), 18189. https://dx.doi.org/10.1038/s41598-019-54539-7)
The authors show that BrfA is an RF (release factor)-dependent ribosome rescue factor in Gram-positive bacteria, such as Bacillus. A custom PUREfrex® kit (lacking E. coli ribosomes and RFs) together with Bacillus subtilis (Bs) ribosomes is used to reveal that the ribosome rescue activity of BrfA acts in conjunction with Bs RF2 but not Bs RF1. (Shimokawa-Chiba, N., Müller, C., Fujiwara, K., Beckert, B., Ito, K., Wilson, D., Chiba, S.(2019). Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis Nature Communications 10(1), 5397. https://dx.doi.org/10.1038/s41467-019-13408-7)
This paper explores the history and progress in creating membrane components for compartmentalization of artificial cells. In one example, PUREfrex® is highlighted for its support of genetically encoded in vesiculo synthesis of GPAT and LPAAT enzymes that act to synthesize phosphatidylethanolamine and phosphatidylglycerol lipids in situ from externally added acyl-CoA and glycerol-3-phosphate. (Vogele, K., Pirzer, T., Simmel, F. (2019). Genetically Encoded Membranes for Bottom‐Up Biology ChemSystemsChem https://dx.doi.org/10.1002/syst.201900016)
Use of a custom PUREfrex® kit to demonstrate the ability to replace the three kinases of the standard PURE system (creatine kinase, myokinase and nucleoside diphosphate kinase) with a single newly characterized bi-functional Cytophaga hutchinsonii polyphosphate kinase, which phosphorylates nucleotides in an exchange reaction from polyphosphate. The new PURE system exhibited yields comparable with unmodified PUREfrex®. (Wang, P., Fujishima, K., Berhanu, S., Kuruma, Y., Jia, T., Khusnutdinova, A., Yakunin, A., McGlynn, S. (2019). A Bifunctional Polyphosphate Kinase Driving the Regeneration of Nucleoside Triphosphate and Reconstituted Cell-Free Protein Synthesis. ACS synthetic biology https://dx.doi.org/10.1021/acssynbio.9b00456)
Use of PUREfrex® to generate artificial cells that exhibit spatiotemporal patterning of Min waves and to study the effect of spatial confinement on IVTT. (Yoshida, A., Kohyama, S., Fujiwara, K., Nishikawa, S., Doi, N.(2019). Regulation of spatiotemporal patterning in artificial cells by a defined protein expression system Chemical Science https://dx.doi.org/10.1039/c9sc02441g).
Comparison of PUREfrex®1, PUREfrex®2 and PURExpress in a method for compartmentalizing PURE system reactions within liposomes and characterizing reactions with high content imaging analysis. (Blanken, D., Nies, P., Danelon, C. (2019). Quantitative imaging of gene-expressing liposomes reveals rare favorable phenotypes Physical Biology 16(4), 045002. https://dx.doi.org/10.1088/1478-3975/ab0c62)
Comparison of PUREfrex®, PUREfrex®2 and PURExpress to identify kinetic parameters and relative efficiency of IVTT. (Doerr, A., Reus, E., Nies, P., Haar, M., Wei, K., Kattan, J., Wahl, A., Danelon, C. (2019). Modelling cell-free RNA and protein synthesis with minimal systems Physical Biology 16(2), 025001.https://dx.doi.org/10.1088/1478-3975/aaf33d)
Use of PUREfrex® to study the role of co-transcriptional folding states (kinetic and thermodynamically favored) and their effect on translational efficiency. (Endoh, T., Sugimoto, N. (2019). Conformational Dynamics of the RNA G-Quadruplex and its Effect on Translation Efficiency Molecules 24(8), 1613. https://dx.doi.org/10.3390/molecules24081613)
Use of PUREfrex® to generate artificial cells expressing a histamine-sensitive aptamer to program three distinct behaviors: expression of mCherry, release of molecular cargo and self-destruction. (Dwidar, M., Seike, Y., Kobori, S., Whitaker, C., Matsuura, T., Yokobayashi, Y. (2019). Programmable Artificial Cells Using Histamine-Responsive Synthetic Riboswitch Journal of the American Chemical Society 141(28), 11103-11114. https://dx.doi.org/10.1021/jacs.9b03300)
Use of PUREfrex® 2.0 to identify optimal conditions for the expression of soluble, properly folded and functional human IgG1, IgG2, and IgG4. This feat required co-expression of two constructs and proper folding and disulfide bond formation of a large tetrameric protein. (Murakami, S., Matsumoto, R., Kanamori, T. (2019). Constructive approach for synthesis of a functional IgG using a reconstituted cell-free protein synthesis system Scientific Reports 9(1), 671. https://dx.doi.org/10.1038/s41598-018-36691-8)
Use of PUREfrex® to measure the proportion of full length and arrested translation products of alpha spectrin, as a means of gleaning folding dynamics. (Kemp, G., Nilsson, O., Tian, P., Best, R., Heijne, G. (2019). Cotranslational folding cooperativity of contiguous domains of α-spectrin bioRxiv https://dx.doi.org/10.1101/653360)
Use of PUREfrex® to perform cell-free protein synthesis in monodisperse picoliter droplets created by centrifugation of an in-microtube step emulsion device. (Shin, D., Morimoto, Y., Sawayama, J., Miura, S., Takeuchi, S. (2019). Centrifuge-based Step Emulsification Device for Simple and Fast Generation of Monodisperse Picoliter Droplets Sensors and Actuators B: Chemical 301(), 127164. https://dx.doi.org/10.1016/j.snb.2019.127164)
Use of PUREfrex® to generate a photosynthesizing artificial cell delimited by giant unilamellar vesicles (GUVs) containing artificial ATP-generating proteoliposomes with membrane-associated bacteriorhodopsin and F°F1ATPase. (Berhanu, S., Ueda, T., Kuruma, Y. (2019). Artificial photosynthetic cell producing energy for protein synthesis Nature Communications 10(1), 1325. https://dx.doi.org/10.1038/s41467-019-09147-4)
Use of PUREfrex® to test if the YcgG2 allele of a stand-alone PDE in neonatal meningitis-causing Escherichia coli (NMEC) strains is an ORF that can be translated into the predicted encoded product. (Zlatkov, N., Uhlin, B.(2019). Absence of Global Stress Regulation in Escherichia coli Promotes Pathoadaptation and Novel c-di-GMP-dependent Metabolic Capability Scientific Reports 9(1), 2600. https://dx.doi.org/10.1038/s41598-019-39580-w)
Use of PUREfrex® to determine the cofactor requirements of ResQ, a gram positive bacterial release factor-dependent ribosome rescue factor. The investigation utilized a B. subtilis hybrid PURE system, a modified version of the PURE coupled transcription-translation system, in which the original E. coli ribosomes were replaced with B. subtilis ribosomes. (Shimokawa-Chiba, N., Müller, C., Fujiwara, K., Beckert, B., Ito, K., Wilson, D., Chiba, S.(2019). ResQ, a release factor-dependent ribosome rescue factor in the Gram-positive bacterium Bacillus subtilis bioRxiv https://dx.doi.org/10.1101/732420)
Use of PUREfrex® to show that the amount of synthesized protein (translation efficiency) depends on a short translational ramp that comprises the first 5 codons in mRNA. Also, the authors show that differences in the short ramp can lead to 3 to 4 orders of magnitude changes in protein abundance. (Verma, M., Choi, J., Cottrell, K., Lavagnino, Z., Thomas, E., Pavlovic-Djuranovic, S., Szczesny, P., Piston, D., Zaher, H., Puglisi, J., Djuranovic, S. (2019). Short translational ramp determines efficiency of protein synthesis bioRxivhttps://dx.doi.org/10.1101/571059)
Use of PUREfrex® to create a self-replicating DNA system in liposome-based artificial cells. (Nies, P., Westerlaken, I., Blanken, D., Salas, M., Mencía, M., Danelon, C. (2018). Self-replication of DNA by its encoded proteins in liposome-based synthetic cells Nature Communications 9(1), 1583.https://dx.doi.org/10.1038/s41467-018-03926-1)
A review including the use of PUREfrex® in the production of stable isotope-labeled internal standards for targeted quantitative proteomics. (Narumi, R., Masuda, K., Tomonaga, T., Adachi, J., Ueda, H., Shimizu, Y.(2018). Cell-free synthesis of stable isotope-labeled internal standards for targeted quantitative proteomics Synthetic and Systems Biotechnology 3(Cell 150 2012), 97-104. https://dx.doi.org/10.1016/j.synbio.2018.02.004)
Use of custom PUREfrex® kits to evaluate the role of folding chaperones on co-translational misassembly of homo- mono and tetrameric proteins. (Natan, E., Endoh, T., Haim-Vilmovsky, L., Flock, T., Chalancon, G., Hopper, J., Kintses, B., Horvath, P., Daruka, L., Fekete, G., Pál, C., Papp, B., Oszi, E., Magyar, Z., Marsh, J., Elcock, A., Babu, M., Robinson, C., Sugimoto, N., Teichmann, S. (2018). Cotranslational protein assembly imposes evolutionary constraints on homomeric proteins Nature Structural & Molecular Biology 25(3), 279-288. https://dx.doi.org/10.1038/s41594-018-0029-5)
Use of PUREfrex® to show the similar folding pathways of an Ig domain (Titin) co-translationally folding in a ribosome versus folding free in solution. (Tian, P., Steward, A., Kudva, R., Su, T., Shilling, P., Nickson, A., Hollins, J., Beckmann, R., Heijne, G., Clarke, J., Best, R. (2018). Folding pathway of an Ig domain is conserved on and off the ribosome Proceedings of the National Academy of Sciences 115(48), 201810523. https://dx.doi.org/10.1073/pnas.1810523115)
Use of PUREfrex® 2.0 to co-translationally express structurally correct, ligand-binding G protein-coupled receptors (CX3CR1 and CCR5) in artificial lipid bilayers and micelles at an efficiency of 0.04-0.1 mg/ml. (Gessesse, B., Nagaike, T., Nagata, K., Shimizu, Y., Ueda, T. (2018). G-Protein Coupled Receptor Protein Synthesis on a Lipid Bilayer Using a Reconstituted Cell-Free Protein Synthesis System Life 8(4), 54. https://dx.doi.org/10.3390/life8040054)
Use of PUREfrex® 2.0 to rapidly evaluate the functional efficiency of a library of bar-coded T7 promoter variants. (Komura, R., Aoki, W., Motone, K., Satomura, A., Ueda, M. (2018). High-throughput evaluation of T7 promoter variants using biased randomization and DNA barcoding PLOS ONE 13(5), e0196905. https://dx.doi.org/10.1371/journal.pone.0196905)
Use of PUREfrex® to express single domain antibodies (Sybodies) that can bind and stabilize membrane proteins in specific conformations. (Zimmermann, I., Egloff, P., Hutter, C., Arnold, F., Stohler, P., Bocquet, N., Hug, M., Huber, S., Siegrist, M., Hetemann, L., Gera, J., Gmür, S., Spies, P., Gygax, D., Geertsma, E., Dawson, R., Seeger, M. (2018). Synthetic single domain antibodies for the conformational trapping of membrane proteins eLife 7, e34317. https://dx.doi.org/10.7554/elife.34317)
Use of PUREfrex® to probe the relationship between the folding of domains in isolation and co-translationally on the ribosome. (Marsden, A., Hollins, J., O'Neill, C., Ryzhov, P., Higson, S., Mendonca, C., Kwan, T., Kwa, L., Steward, A., Clarke, J. (2018). Investigating the Effect of Chain Connectivity on the Folding of a Beta-Sheet Protein on and off the Ribosome Journal of Molecular Biology 430(Curr. Opin. Struct. Biol. 42 2017), 5207-5216. https://dx.doi.org/10.1016/j.jmb.2018.10.011)
Use of PUREfrex® 1.0 to examine whether cytosolic factors contribute to the elongation arrest of MifM protein. (Fujiwara, K., Ito, K., Chiba, S. (2018). MifM-instructed translation arrest involves nascent chain interactions with the exterior as well as the interior of the ribosome Scientific Reports 8(1), 10311. https://dx.doi.org/10.1038/s41598-018-28628-y)
Use of PUREfrex® to study the role of bacterial Hsp70 chaperone ortholog DnaK in curli biogenesis and biofilm formation. The study shows the use of DnaK mix to demonstrate the direct requirement of DnaK, DnaJ and GrpE to promote proper folding and function of PUREfrex®-synthesized CsgD protein. (Sugimoto, S., Arita-Morioka, K., Terao, A., Yamanaka, K., Ogura, T., Mizunoe, Y. (2018). Multitasking of Hsp70 chaperone in the biogenesis of bacterial functional amyloids Communications Biology 1(1)https://dx.doi.org/10.1038/s42003-018-0056-0)
Use of PUREfrex® ribosomes to induce human dermal fibroblast transdifferentiation into multipotent cell clusters. (Ito, N., Katoh, K., Kushige, H., Saito, Y., Umemoto, T., Matsuzaki, Y., Kiyonari, H., Kobayashi, D., Soga, M., Era, T., Araki, N., Furuta, Y., Suda, T., Kida, Y., Ohta, K. (2018). Ribosome Incorporation into Somatic Cells Promotes Lineage Transdifferentiation towards Multipotency Scientific Reports 8(1), 1634. https://dx.doi.org/10.1038/s41598-018-20057-1)
Use of PUREfrex® to reveal a novel cellular Mg2+ sensing mechanism that exploits abortive translation caused by Intrinsic Ribosome Destabilization (IRD) triggered by N-terminal acidic and proline intermitted sequences. (Chadani, Y., Niwa, T., Izumi, T., Sugata, N., Nagao, A., Suzuki, T., Chiba, S., Ito, K., Taguchi, H.(2017). Intrinsic Ribosome Destabilization Underlies Translation and Provides an Organism with a Strategy of Environmental Sensing Molecular Cell 68(3), 528-539.e5. https://dx.doi.org/10.1016/j.molcel.2017.10.020)
Use of PUREfrex® to express novel library genes for functional analysis. (Judd, J., Boucher, N., Roey, E., Gray, T., Derbyshire, K. (2017). Application of Distributive Conjugal DNA Transfer in Mycobacterium smegmatis To Establish a Genome-Wide Synthetic Genetic Array Journal of Bacteriology 199(20), e00410-17. https://dx.doi.org/10.1128/jb.00410-17)
Use of PUREfrex® to express proteins for analysis in a biotin-polyethylene glycol (PEG)-conjugated maleimide (biotin-PEG-MAL) labeling gel shift assay. Use of an E. coli cysteinyl-tRNA synthetase (EcCARS)-deficient custom PUREfrex® kit to assess the protein synthesis potential of mutant versions of EcCARS. (Akaike, T., Ida, T., Wei, F., Nishida, M., Kumagai, Y., Alam, M., Ihara, H., Sawa, T., Matsunaga, T., Kasamatsu, S., Nishimura, A., Morita, M., Tomizawa, K., Nishimura, A., Watanabe, S., Inaba, K., Shima, H., Tanuma, N., Jung, M., Fujii, S., Watanabe, Y., Ohmuraya, M., Nagy, P., Feelisch, M., Fukuto, J., Motohashi, H. (2017). Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics Nature Communications 8(1), 1177. https://dx.doi.org/10.1038/s41467-017-01311-y)
Use of PUREfrex® to rapidly express a library of site saturation reverse transcriptase mutants for subsequent selection of variants optimized for thermostability, a property difficult to screen for in intact organisms. (Katano, Y., Li, T., Baba, M., Nakamura, M., Ito, M., Kojima, K., Takita, T., Yasukawa, K. (2017). Generation of thermostable Moloney murine leukemia virus reverse transcriptase variants using site saturation mutagenesis library and cell-free protein expression system Bioscience, Biotechnology, and Biochemistry 81(12), 1-7. https://dx.doi.org/10.1080/09168451.2017.1394790)
Use of PUREfrex® to elucidate the effects of different stop codons on the translational termination inhibition efficiency of anti-microbial peptide apidaecin. Apidaecin is shown to act by binding to the A site of ribosomes translating mRNAs terminated with UAG codons. (Matsumoto, K., Yamazaki, K., Kawakami, S., Miyoshi, D., Ooi, T., Hashimoto, S., Taguchi, S. (2017). In vivo target exploration of apidaecin based on Acquired Resistance induced by Gene Overexpression (ARGO assay) Scientific Reports 7(1), 12136.https://dx.doi.org/10.1038/s41598-017-12039-6)
Use of PUREfrex® to study the role of ribosomal protein L31 in different ribosome preparations to promote translation of a DHFR-encoding template. (Ueta, M., Wada, C., Bessho, Y., Maeda, M., Wada, A. (2017). Ribosomal protein L31 in Escherichia coli contributes to ribosome subunit association and translation, whereas short L31 cleaved by protease 7 reduces both activities Genes to Cells 22(5), 452-471. https://dx.doi.org/10.1111/gtc.12488)
Comparison of the performance of PURE (PUREfrex®) and E coli extract-based in vitro transcription/translation systems. Reliance of this study on protein activity (GFP fluorescence) without regard to full length protein yields renders the analysis rather crude and limited. (Reyes, S., Kuruma, Y., Tsuda, S.(2017). Uncovering cell-free protein expression dynamics by a promoter library with diverse strengths bioRxiv https://dx.doi.org/10.1101/214593)
Use of PUREfrex® reactions performed within liposomes to generate proteoliposomes harboring membrane-associated lipid-synthesizing enzymes capable of generating and incorporating phospholipids from precursors G3P and acyl-CoA. Protein synthesis and lipid biogenesis, two essential functions for cell homeostasis, occurred in a single environment demonstrating the compatibility of these biological modules for further integration into a semi-synthetic minimal cell. (Scott, A., Noga, M., Graaf, P., Westerlaken, I., Yildirim, E., Danelon, C. (2016). Cell-Free Phospholipid Biosynthesis by Gene-Encoded Enzymes Reconstituted in Liposomes PLOS ONE 11(10), e0163058. https://dx.doi.org/10.1371/journal.pone.0163058)
Use of PUREfrex® to produce ADAMTS4 and ADAMTS5 domain-specific fragments for immunoreactivity analyses. (Shiraishi, A., Mochizuki, S., Miyakoshi, A., Kojoh, K., Okada, Y. (2016). Development of human neutralizing antibody to ADAMTS4 (aggrecanase-1) and ADAMTS5 (aggrecanase-2) Biochemical and Biophysical Research Communications 469(1), 62-69. https://dx.doi.org/10.1016/j.bbrc.2015.11.072)
Use of PUREfrex® to demonstrate the translational inhibitory effect on FlaA expression of post-transcriptional regulator CsrA. (Radomska, K., Ordoñez, S., Wösten, M., Wagenaar, J., Putten, J. (2016). Feedback control of Campylobacter jejuni flagellin levels through reciprocal binding of FliW to flagellin and the global regulator CsrA Molecular Microbiology 102(2), 207-220. https://dx.doi.org/10.1111/mmi.13455)
Use of PUREfrex® to study translational pausing, comparing synthesis in vitro and in vivo in parallel to illuminate translation as an elementary process carried out by the basic translation machinery (in vitro) as well as to profile translation orchestrated with cellular factors (in vivo). (Chadani, Y., Niwa, T., Chiba, S., Taguchi, H., Ito, K. (2016). Integrated in vivo and in vitro nascent chain profiling reveals widespread translational pausing Proceedings of the National Academy of Sciences 113(7), E829-E838. https://dx.doi.org/10.1073/pnas.1520560113)
Use of PUREfrex® to express bi-specific antibody fragments (diabodies, scFvs and taFvs) and to select for high affinity binders using cell free DNA display technology. (Nakayama, M., Komiya, S., Fujiwara, K., Horisawa, K., Doi, N. (2016). In vitro selection of bispecific diabody fragments using covalent bicistronic DNA display Biochemical and Biophysical Research Communications 478(2), 606-611. https://dx.doi.org/10.1016/j.bbrc.2016.07.113)
Use of PUREfrex® to study the role of chaperones (TF: Trigger Factor; and GroEL/ES) on modulating cotranslational protein folding forces and their effects on translation rate. TF, but not GroEL/ES, is shown to strongly reduce the force generated by the folding of DHFR (which folds after leaving the ribosomal exit tunnel). By contrast neither chaperone affects the force generated by the folding of ADR1a (that folds within the exit tunnel). (Nilsson, O., Müller-Lucks, A., Kramer, G., Bukau, B., Heijne, G. (2016). Trigger Factor Reduces the Force Exerted on the Nascent Chain by a Cotranslationally Folding Protein Journal of Molecular Biology 428(6), 1356-1364. https://dx.doi.org/10.1016/j.jmb.2016.02.014)
Use of PUREfrex® to demonstrate the utility of liposomes to promote the expression, proper membrane incorporation and channel function of the potassium transporter KcsA. The authors conclude: The cell-free protein synthesis/liposome system is a promising method to achieve high throughput analysis of channel proteins. (Ando, M., Akiyama, M., Okuno, D., Hirano, M., Ide, T., Sawada, S., Sasaki, Y., Akiyoshi, K. (2015). Liposome chaperon in cell-free membrane protein synthesis: one-step preparation of KcsA-integrated liposomes and electrophysiological analysis by the planar bilayer method Biomaterials Science 4(2), 258-264. https://dx.doi.org/10.1039/c5bm00285k)
Use of PUREfrex® to evaluate the effects of liposomes on folding of 85 aggregation-prone membrane proteins from Escherichia coli. The reconstituted cell-free translation system circumvented the problem of debris that is often present in other translation systems, permitting unobstructed direct analysis of liposome effects. (Niwa, T., Sasaki, Y., Uemura, E., Nakamura, S., Akiyama, M., Ando, M., Sawada, S., Mukai, S., Ueda, T., Taguchi, H., Akiyoshi, K. (2015). Comprehensive study of liposome-assisted synthesis of membrane proteins using a reconstituted cell-free translation system Scientific Reports 5(1), 18025. https://dx.doi.org/10.1038/srep18025)
Use of PUREfrex® to investigate intrinsic features of recombinant proteins without contamination from the activity of other intracellular components such as heat shock chaperones. (Yamamoto, H., Shima, T., Yamaguchi, M., Mochizuki, Y., Hoshida, H., Kakuta, S., Kondo-Kakuta, C., Noda, N., Inagaki, F., Itoh, T., Akada, R., Ohsumi, Y. (2015). The Thermotolerant Yeast Kluyveromyces marxianus Is a Useful Organism for Structural and Biochemical Studies of Autophagy Journal of Biological Chemistry 290(49), 29506-29518. https://dx.doi.org/10.1074/jbc.m115.684233)
Use of PUREfrex® coupled with Vibrio alginolyticus ribosomes to show that the regulatory protein, VemP, is synthesized mainly as a peptidyl-tRNA arrested near its C terminus. Regulation of this arrest influences the availability of a downstream Shine Delgarno sequence governing the translation of SecDF2. The results showed that VemP-mediated regulation of SecDF2 is essential for the survival of the marine bacterium V. alginolyticus in low-salinity environments. (Ishii, E., Chiba, S., Hashimoto, N., Kojima, S., Homma, M., Ito, K., Akiyama, Y., Mori, H. (2015). Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria Proceedings of the National Academy of Sciences 112(40), E5513-E5522. https://dx.doi.org/10.1073/pnas.1513001112)
Use of PUREfrex® to perform in vitro arrest peptide (AP) assays with constructs designed to measure the cotranslational folding-related force generated by Adr1a on the ribosome. The results show that cotranslational folding for a small domain protein like Adr1a can occur within the exit tunnel of the ribosome. (Nilsson, O., Hedman, R., Marino, J., Wickles, S., Bischoff, L., Johansson, M., Müller-Lucks, A., Trovato, F., Puglisi, J., O’Brien, E., Beckmann, R., von Heijne, G. (2015). Cotranslational Protein Folding inside the Ribosome Exit Tunnel Cell Reports 12(10), 1533-1540. https://dx.doi.org/10.1016/j.celrep.2015.07.065)
Use of PUREfrex® to probe the ability of SecM, a bacterial secretion monitor protein, to arrest its own protein synthesis. This paper provides evidence supporting the idea that interactions between the nascent chain and the outside of the ribosome contribute to the stability of translational arrest. (Yang, Z., Iizuka, R., Funatsu, T.(2015). Nascent SecM Chain Outside the Ribosome Reinforces Translation Arrest PLOS ONE 10(3), e0122017. https://dx.doi.org/10.1371/journal.pone.0122017)
Articles citing PURExpress®
Use of PURE system (PURExpress®) in a high-density femtoliter droplet array (FEMdA), consisting of 1 million uniform droplets per 1 cm2 to carry out high-throughput protein synthesis and screening. (Zhang, Y., Minagawa, Y., Kizoe, H., Miyazaki, K., Iino, R., Ueno, H., Tabata, K., Shimane, Y., Noji, H. (2019). Accurate high-throughput screening based on digital protein synthesis in a massively parallel femtoliter droplet array Science Advances 5(8), eaav8185. https://dx.doi.org/10.1126/sciadv.aav8185)
This study introduces ROSALIND (RNA output sensors activated by ligand induction), a fluorescent aptamer-based biosensor system capable of aqueous detection of non-nucleic acid targets such as antibiotics, metals and other small molecules. Allosteric transcription factors regulate the in vitro transcription of a fluorescence-activating RNA aptamer. Fluorescence-activation from aptamer transcription reactions is found to be faster and brighter than fluorescence from transcription-translation (PURE) reactions of sfGFP. (Alam, K., Jung, J., Verosloff, M., Clauer, P., Lee, J., Capdevila, D., Pastén, P., Giedroc, D., Collins, J., Lucks, J. (2019). Rapid, Low-Cost Detection of Water Contaminants Using Regulated In Vitro Transcription bioRxiv https://dx.doi.org/10.1101/619296)
PURExpress® was used in the development of low cost, abiotic, paper-based, freeze-dried cell-free transcription/translation sensors to rapidly and species-specifically quantify bacterial and host RNAs from stool samples. The method’s utility is demonstrated in the discrimination of active and passive Clostridium difficile infection by the quantitation of toxin mRNA. (Takahashi, M., Tan, X., Dy, A., Braff, D., Akana, R., Furuta, Y., Donghia, N., Ananthakrishnan, A., Collins, J. (2018). A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers Nature Communications 9(1), 3347. https://dx.doi.org/10.1038/s41467-018-05864-4)
PURExpress® was used in the development and validation of a low cost, abiotic, paper-based, freeze-dried cell-free transcription/translation sensor for the detection and discrimination of Norovirus. It employs an initial isothermal RT-RPA or NASBA step that enables detection down to 270 aM and, if employing an additional synbody-mediated viral particle enrichment step, 270 zM. (Ma, D., Shen, L., Wu, K., Diehnelt, C., Green, A.(2018). Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment Synthetic Biology 3(1), ysy018-. https://dx.doi.org/10.1093/synbio/ysy018)
Use of PURExpress® to study variation in cell free expression systems including in various single and two gene circuits (Chizzolini, F., Forlin, M., Martín, N., Berloffa, G., Cecchi, D., Mansy, S. (2017). Cell-free translation is more variable than transcription ACS Synthetic Biology 6(4), 638 647. https://dx.doi.org/10.1021/acssynbio.6b00250)
Use of PURExpress® to dissect the limiting factors of PURE system-based protein synthesis as compared to lysate-based protein synthesis. This provides a good comparison of PURE versus lysate-based reaction performance. Since this paper was published PUREfrex® has evolved significantly in addressing protein folding and functionality by including supplements that promote the formation of correct folding and disulfide bond formation. It would be interesting to see an updated side by side comparison of the two types of cell free protein synthesis systems. (Li, J., Zhang, C., Huang, P., Kuru, E., Forster-Benson, E., Li, T., Church, G. (2017). Dissecting limiting factors of the Protein synthesis Using Recombinant Elements (PURE) systemTranslation 5(1), e1327006. https://dx.doi.org/10.1080/21690731.2017.1327006)
PURExpress® was used in the development and validation of low cost, abiotic, freeze-dried, cell-free transcription/translation, paper-based sensors for the detection and discrimination of Zika and related viral RNA species. This was achieved by linking isothermal RNA amplification (in NASBA reactions) to toehold switch RNA sensors. A CRISPR/Cas9-based module added the ability to discriminate related viral strains by a single base difference. (Pardee, K., Green, A., Takahashi, M., Braff, D., Lambert, G., Lee, J., Ferrante, T., Ma, D., Donghia, N., Fan, M., Daringer, N., Bosch, I., Dudley, D., O’Connor, D., Gehrke, L., Collins, J. (2016). Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components Cell 165(5), 1255-1266. https://dx.doi.org/10.1016/j.cell.2016.04.059)
PURExpress® was used as a control to show that a prokaryotic PURE system could translate circular RNA templates. (Abe, N., Matsumoto, K., Nishihara, M., Nakano, Y., Shibata, A., Maruyama, H., Shuto, S., Matsuda, A., Yoshida, M., Ito, Y., Abe, H. (2015). Rolling Circle Translation of Circular RNA in Living Human Cells. Scientific reports 5(1), 16435. https://dx.doi.org/10.1038/srep16435)
Articles citing PURESYSTEM®
Use of PURESYSTEM® (BioComber, available from Cosmobio USA) and polyethylene glycol (PEG)-charged amino phenylalanine amber tRNAs to incorporate PEG into specific sites in Thioredoxin (Trx) and a small ProX peptide tag. (Tada, S., Andou, T., Suzuki, T., Dohmae, N., Kobatake, E., Ito, Y. (2012). Genetic PEGylationPLoS ONE 7(11), e49235 6. https://dx.doi.org/10.1371/journal.pone.0049235)
Articles using home-made PURE systems
A method is presented to easily produce a low cost home-made OnePot PURE system, capable of protein yields of 156ug/mL at a cost of 0.09 USD/uL (compared to 1.36 USD/uL for the commercial PURExpress® kit from NEB). (Lavickova, B., Maerkl, S. (2018). A simple, robust, and low-cost method to produce the PURE cell-free system bioRxiv https://dx.doi.org/10.1101/420570)
This paper introduces a species-independent translational leader sequence (SITS) that bypasses the requirement for 5’-capped RNA and translation initiation factors, thereby permitting the comparison of transcriptional templates across diverse eukaryotic and prokaryotic extract-based cell free expression systems. Further, a new cell-free extract system based on the Trypanosomatidae parasite Leishmania tarentolae is presented. Coupled with SITS, this system has the following advantages: amenable to large-scale and inexpensive fermentation culture, genetic manipulation, capable of co-expressing multiple proteins and compatible with direct spectroscopic analysis of protein:protein and protein:small molecule interaction. (Mureev, S., Kovtun, O., Nguyen, U., Alexandrov, K. (2009). Species-independent translational leaders facilitate cell-free expression Nature Biotechnology 27(8), 747-752. https://dx.doi.org/10.1038/nbt.1556)
This paper provides mechanistic insight into the E. coli lysate-based Cytomim cell-free protein synthesis system. This cost-effective system (using pyruvate as an energy source) is shown to co-activate central metabolism, oxidative phosphorylation, transcription, translation and protein folding. (Jewett, M., Calhoun, K., Voloshin, A., Wuu, J., Swartz, J. (2008). An integrated cell‐free metabolic platform for protein production and synthetic biology Molecular Systems Biology 4(1), 220. https://dx.doi.org/10.1038/msb.2008.57)
Articles describing cell-free protein synthesis applications
An up to date and comprehensive review focused on applications for cell free extract-based expression systems. Applications range from gene expression in non-model organisms, artificial cells, genetic networks to on-demand biosensing, biomanufacturing, and synthetic biology education. (Silverman, A., Karim, A., Jewett, M. (2019). Cell-free gene expression: an expanded repertoire of applications Nature Reviews Genetics https://dx.doi.org/10.1038/s41576-019-0186-3)
This study introduces a method to extend the range of molecules that can be detected by cell-free transcription/translation biosensors. The authors expand on their SensiPath software package that identifies catabolic cascades that can convert undetectable molecules into ones that can be detected by existing allosteric transcription factors. They provide proof of principle results with detection of benzoic acid, hippurate and cocaine in real world contexts. (Delépine, B., Libis, V., Carbonell, P., Faulon, J. (2016). SensiPath: computer-aided design of sensing-enabling metabolic pathways Nucleic Acids Research 44(W1), W226-W231. https://dx.doi.org/10.1093/nar/gkw305)
This study provides yet another example of a cell-free transcription/translation biosensor system that uses a cocktail of three catabolic enzymes to convert atrazine (an herbicide and suspected endocrine-disrupting compound) into a molecule that can be directly sensed with the well-characterized allosteric transcription factor AtzR. (Silverman, A., Akova, U., Alam, K., Jewett, M., Lucks, J. (2019). Design and optimization of a cell-free atrazine biosensor bioRxiv https://dx.doi.org/10.1101/779827)
Cell-free transcription/translation biosensor systems are challenged by matrix-specific interference inherent in complex samples such as blood and urine. This paper introduces a generalizable parallel calibration strategy that uses the sample matrix itself to generate custom reference curves to permit quantitative diagnostics. (McNerney, M., Zhang, Y., Steppe, P., Silverman, A., Jewett, M., Styczynski, M. (2019). Point-of-care biomarker quantification enabled by sample-specific calibration Science Advances 5(9), eaax4473. https://dx.doi.org/10.1126/sciadv.aax4473)
An up to date review of efforts to engineer the translation machinery itself (AARSs/tRNAs, initiation factors, elongation factors, termination factors, ribosomes) to optimize non-natural amino acid incorporation in cell-free in vitro transcription/translation reactions using extract-based and PURE system-based approaches. (Hammerling, M., Krüger, A., Jewett, M. (2019). Strategies for in vitro engineering of the translation machinery Nucleic Acids Research https://dx.doi.org/10.1093/nar/gkz1011)
Modular cell-free bacterial extract- and PURE system-based freeze-dried protein expression reactions are introduced as a new paradigm for bringing affordable life sciences and biotechnology experiments into any classroom, making quality biology education accessible to all students. Modules are presented to teach tunable protein expression, enzymatic reactions, biomaterial formation, and biosensors using RNA switches. (Huang, A., Nguyen, P., Stark, J., Takahashi, M., Donghia, N., Ferrante, T., Dy, A., Hsu, K., Dubner, R., Pardee, K., Jewett, M., Collins, J. (2018). BioBits™ Explorer: A modular synthetic biology education kitScience Advances 4(8), eaat5105. https://dx.doi.org/10.1126/sciadv.aat5105)
A review of cell free protein synthesis systems from the advent to now, highlighting supplements added to these systems and their effects. (Dopp, B., Tamiev, D., Reuel, N. (2018). Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract Biotechnology Advances 37 (Biochem. Biophys. Res. Commun. 338 2005), 246-258. https://dx.doi.org/10.1016/j.biotechadv.2018.12.006)
This is a proof of principle study introducing a novel biosensor for quorum sensing molecules in P. aeruginosa-infected respiratory samples. The system comprises an E. coli lysate-based cell-free transcription/translation module to expresses a plasmid encoding the P. aeruginosa acyl homoserine lactone (AHL)-detecting transcriptional inducer LasRV and a downstream LasRV-responsive promoter driving GFP expression. AHL binding by LasRV allosterically induces its ability to activate GFP transcription. (Wen, K., Cameron, L., Chappell, J., Jensen, K., Bell, D., Kelwick, R., Kopniczky, M., Davies, J., Filloux, A., Freemont, P.(2017). A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P. aeruginosa-Infected Respiratory Samples ACS Synthetic Biology 6(12), 2293-2301. https://dx.doi.org/10.1021/acssynbio.7b00219)
This report introduces a lyophilized, field use-compatible biosensor system for endocrine disrupting chemicals (EDCs). Called RAPID (Rapid Adaptable Portable In-vitro Detection), the system comprises a homemade E. coli lysate-based cell-free protein synthesis module that expresses an engineered, allosterically-activated fusion protein, which contains the ligand binding domain from a target nuclear hormone receptor. The system can detect EDC presence or absence in environmental samples and could also be applied for drug screening. (Salehi, A., Tang, M., Smith, M., Hunt, J., Wood, D., Bundy, B. (2017). A Cell-free Protein Synthesis Approach to Biosensing hTRB-Specific Endocrine Disruptors Analytical Chemistry https://dx.doi.org/10.1021/acs.analchem.6b04034)
Demonstration of the construction of a set of 3 plasmids encoding 30 translation factors to facilitate scale up of expression and purification of PURE system components. (Shepherd, T., Du, L., Liljeruhm, J., Samudyata, Wang, J., Sjödin, M., Wetterhall, M., Yomo, T., Forster, A. (2017). De novo design and synthesis of a 30-cistron translation-factor module Nucleic Acids Research 45(18), gkx753-. https://dx.doi.org/10.1093/nar/gkx753)
A review of cell free protein synthesis focusing on the pros and cons of pro- versus eukaryotic lysate-based systems. (Zemella, A., Thoring, L., Hoffmeister, C., Kubick, S. (2015). Cell-Free Protein Synthesis: Pros and Cons of Prokaryotic and Eukaryotic Systems ChemBioChem 16(17), 2420 2431. https://dx.doi.org/10.1002/cbic.201500340)
A review of cell-free protein synthesis focused on the PURESYSTEM from BioComber (available from Cosmobio USA). (Whittaker, J. (2012). Cell-free protein synthesis: the state of the art Biotechnology letters 35(2), 143 152. https://dx.doi.org/10.1007/s10529-012-1075-4)
A dated but still useful review of the early history of cell-free protein synthesis leading to Ueda’s PURE system. (Chong, S. (2001). Overview of Cell-Free Protein Synthesis: Historic Landmarks, Commercial Systems, and Expanding Applications Current Protocols in Molecular Biology 101(1), 16.30.1 16.30.11. https://dx.doi.org/10.1002/0471142727.mb1630s108)

